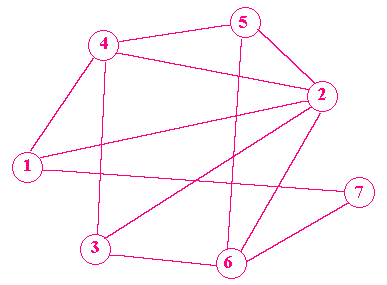
The Gene MachineToby HowardThis article first appeared in Personal Computer World magazine, December 1996 AS RESEARCHERS CONTINUE to look beyond silicon for the computers of the future, a US scientist has created a massively parallel computer in a single test-tube containing a few drops of liquid. His computer is DNA, the molecule of life. Leonard Adleman, a computer scientist at the University of Southern California, has devised a novel solution to a classic mathematical problem. In doing so, he has single-handedly laid the foundation for a new technology. Imagine that a travelling salesman has to visit a number of towns, starting at one specified town and finishing in another. Given that some roads between towns may allow only one-way travel, and that not all towns will have direct roads between them, is it possible for the salesman to find a route
such that he visits each town once only, in a continuous path? The diagram shows the test case used by Adleman. Here, there are seven towns, 1 to 7, and the arrows between towns show the interconnecting roads and the allowed direction of travel. The salesman must start in town 1 and finish in town 7. Such a simple case can be solved with a few minutes' trial and error (the answer is 1 -> 2 -> 3 -> 4 -> 5 -> 6 -> 7) but as the number of towns and their interconnections increase, the problem becomes very hard to solve indeed. In fact, the problem belongs to one of the hardest classes of mathematical problems known, which require enormous computing power to attack. Adleman's breakthrough was to use DNA to solve the problem, and his approach was ingenious. This is how he did it. DNA comprises two intertwined molecular strands, each of which is a long chain of alternating phosphates and sugars. Attached to each sugar is a molecular group called a 'base', and there are four different kinds, known as A, C, G and T. It is the particular sequence of bases along a strand that forms the genetic code for life. An A base on one strand attacts a T base on the other strand, and a C base attracts a G base. These attractions pull the two strands together into the familiar 'double helix' shape discovered by Watson and Crick in 1953. Adleman represented each city, and each road between two cities, with a specially engineered strand of DNA exactly 20 bases long. The sequence of bases in each strand was carefully designed such that strands could link with each other to spell out possible routes. Take, for example, cities 6 and 2. The strand of DNA representing the road from 6 to 2 would stick to the end of the strand representing city 6, and the beginning of the strand for city 2, but not to any part of any strands for other cities. To solve the problem of finding a route beween cities 1 and 7, Adleman mixed together in his test-tube a million million copies of all the possible strands for the cities and their interconnections, and allowed them to link up with each other. Next, he used standard biochemical techniques to isolate particular strands. First, he isolated only those linked-up strands which started with the code for 1, and ended with 7. Then, he isolated only those strands which coded a route through seven cities, knowing that these strands must be exactly 140 (7 x 20) bases long. Longer or shorter strands were rejected. Finally, he kept only those strands containing city 1, and of these he kept only those containing city 2, and so on. After seven days of intensive laboratory work, Adleman's test-tube contained the answer to the problem, subsequently visible as a series of dark bands on a DNA sequencing gel. On the face of it, it might hardly seem worth the bother, especially as Adleman already knew the answer before he started the experiment. But this was much more than a curious laboratory stunt. During the initial 'linking-up' stage of the process, Adleman's test-tube computer effectively performed an astonishing 10^14 calculations. And it did so with the consumption of only a tiny amount of energy, and in a tiny physical space. This was the first time that the combinatorial power of DNA had ever been exploited for computation, and Adleman's work has sparked a flurry of activity. The first researcher to take the idea further was Richard Lipton of Princeton University, who showed how to use DNA to solve another important puzzle in computer science: the 'satifiability' problem, routinely faced by designers of logic circuits. Here, the goal is to find the solutions to problems in Boolean logic. For example, given an expression such as: ( (a = 1) OR (b = 1) OR (c = 0) ) AND ( (b = 0) OR (c = 1) ) the problem is to find which (if any) binary values of a, b, and c satify the expression. Like the travelling salesman problem, simple instances are easy to solve by trial and error, but as the number of variables and constraints increase, the computation time mushrooms exponentially and the problem soon becomes intractable. With DNA strands, however, huge numbers of potential solutions can be evaluated and discarded in parallel, until the correct solution, if there is one, remains. Perhaps the most exciting proposal is Lipton's scheme for using DNA to code arbitrary binary numbers, which opens up the possibility of DNA-based solutions to a wider range of problems, such as matrix manipulation, factoring, dynamic linear programming and algebraic symbol processing. Since the methods of DNA computing are quite different from traditional step-by-step algorithms, perhaps we shall see the development of hybrid machines, part silicon and part DNA. Another promising application is to provide pure data storage: to encode one bit of data using DNA would occupy approximately 1 cubic nanometre, which means a test-tube ought to comfortably accommodate several hundred million gigabytes. Think what you could do with a bathful. Research into DNA computing is taking off in a big way. This year the 2nd Annual Workshop on DNA-based computing was held at Princeton University, and there is already a new scientific journal devoted to the subject. However, like many ideas for computers based on technologies other than silicon, although the DNA computer looks great on paper, the practical biochemical and engineering challenges are immense. DNA manipulations involve fearsomely complicated lab protocols, and are highly prone to contamination and error. Some scientists also warn of the potential ecological horrors of flushing discarded DNA computers down the drain. But apart from the technological excitement, all this talk about using DNA for computing has got the philosophers hopping too. Are the processes inside our own cells essentially performing computations to which human life is the answer? If this is so, the philosophers ask, then what is the question? For more on DNA computing, visit www.ecl.udel.edu/~magee/. Toby Howard teaches at the University of Manchester. |